
 |
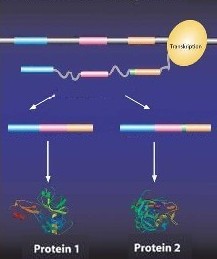
| One gene – two different proteins. The second "NAG" in a tandem cleavage site "NAGNAG" has been colored green. (grafic: InformationsSekretariat A Biotechnologie, modified) |
A human being does not have many more genes than a worm does. But human genes can produce many more proteins than a worm does. How does this work? Now NGFN researchers have been able to trace one of these mechanisms.
Our genes are built up like a mosaic. Between areas containing important information for the production of a protein, there are sections without blueprint information whose functions are still largely unknown.
For the production of a protein, a continuous copy of the gene is made. For the final blueprint, all areas without any information are cut out. The mechanism of cutting out, which occurs in all cells of our body, is called “splicing” in medical terminology. In order for the proper parts of the blueprint to be assembled, there is a marking at the sites of areas with and without blueprints. It is a recognition code of sorts which is labeled “NAG”.
Scientists from the National Genome Research Network (NGFN) and from the Center for Bioinformatics in Jena examined 20,000 genes and found out that in more than 8,000 genes, this short recognition code occurs twice, back-to-back. It is a tandem: “NAGNAG”.
Is this “on purpose” or is it a coincidence? And which of the two markings does the cell use when trimming the protein blueprint?
In more than 800 of these 8,000 genes, there are already experimental clues that it is sometimes the first, but alternatively also the second NAG marking which is used. Thus two product variants, respectively, come into existence: a protein following a blueprint from which all NAG markings have been cut out, and an altered protein following another blueprint, whose second NAG-recognition code ends up in the protein blueprint.
The two protein variants differ from each other only marginally. Sometimes a protein component is added or it disappears; sometimes a protein building block is exchanged for two other ones. However, it is not left to chance how splicing-recognition sites are spliced together. The scientists could show that in some tissues of the body, only one of the two markings is used.
Such minor alterations could also be medically significant. "Dysfunctions with this form of alternative splicing could be the cause of complex diseases in humans, which are caused by several genes," said Dr. Matthias Platzer, who is head of the research team. "What is interesting is that we could find such tandem splicing areas in genes whose mutations cause obesity. Now we will examine whether diseases like obesity, chronic inflammation, Alzheimer’s etc. can be connected to dysfunctions of the structure and function of tandem recognition sites. Perhaps this could be a target for new therapy concepts.”
... further information:




